My research will help biomedical researchers make better use of biomedical data, enabling them to more easily make connections between a variety of domains, including identifying new disease targets for existing drugs and connecting clinical observations with molecular mechanisms.
There is a growing quantity of biomedical data becoming publicly available, but these data are often difficult to utilize in conjunction with each other due to having a wide variety of representational formats. As a part of the National Center for Advancing Translational Science (NCATS) Biomedical Translator program, I and a team consisting of members from OSU, Penn State, and Institute of Systems Biology are developing components of an automated reasoning system to help translational researchers explore existing data in a normalized semantic space. This work would not be possible without the help of my mentor, Dr. Stephen Ramsey.
An example query that the system can answer is something along the lines of “Which approved drugs directly target proteins related to parkinson’s disease?”. This kind of tool helps medical researchers develop and explore hypothesis, come up with novel purposes for existing treatments, and personalize care for patients with rare diseases.
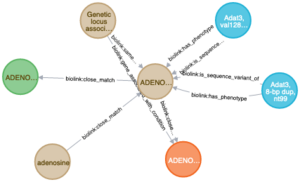
My specific contributions to the project have been to constructing a large knowledge graph to serve as a knowledge basis for the reasoning system. A knowledge graph is just a specific way of representing data: nodes represent entities like proteins, drugs, and diseases, and edges represent the interactions and relationships between them. Since the databases the knowledge graph are continuously being updated, it's important that our build system is able to reingest data on a semi regular basis.
The knowledge graph build system ingests a variety of data sources and ontologies from flat files in a variety of formats and converts them into a standardized knowledge graph that follows the Biolink model. The Biolink model is an hierarchical vocabulary that allows flexible expression of multiple terminologies. Because remapping terms from one terminologies is sometimes an imprecise art, the original terminology is preserved to provide additional context for each node and edge. The purpose of the knowledge graph is to serve as a knowledge base for a reasoning system to use to help choose answers to the biomedical queries with graph traversal metrics and machine learning models that assist in ranking the results.

